|
|
BIOINFORMATICS RESEARCH LABORATORY (BRL) |
“Bioinformatics for cell, molecular and synthetic biology”Dr. Vasilis J. Promponas, PhD is an Associate Professor in Bioinformatics and head of the Bioinformatics Research Laboratory (BRL) at the Department of Biological Sciences, University of Cyprus, Nicosia, Cyprus. He holds a BSc in Physics (Univ. of Athens, 1996) and a PhD in Biological Sciences (Univ. of Athens, 2004). He received an EMBL-EBI visitors program fellowship (1998) and conducted PostDoctoral research at the Department of Cell Biology and Biophysics (Univ. of Athens, 2004-2005). In 2005 he established the BRL (Univ. of Cyprus). Current BRL members (1 Assoc. Prof., 1 PostDoc, 1 PhD student, 2 MSc students and 5 final year undergraduate students) come from diverse backgrounds (Biology, Physics, Bioengineering). Dr Promponas’ work has been published in >60 international peer reviewed journal papers, as well as in the proceedings of international conferences and edited volumes (citations: >16000; h-index:25; source: Google Scholar 09/2024). He serves as Academic Editor for PLOS One (Public Library of Science) and Computational & Structural Biotechnology (Elsevier) and regularly reviews for major international journals publishing research in bioinformatics, computational biology and genomics (e.g. Briefings in Bioinformatics, Bioinformatics, Nucleic Acids Research, NAR Genomics and Bioinformatics, Genome Research, PLOS Computational Biology). Currently, he is a member of the steering committee of the ELIXIR-Cyprus node and an active member of the ELIXIR Intrinsically Disordered Proteins Community. Associate Professor Vasilis J. Promponas Bioinformatics Research Laboratory Department of Biological Sciences; University of Cyprus
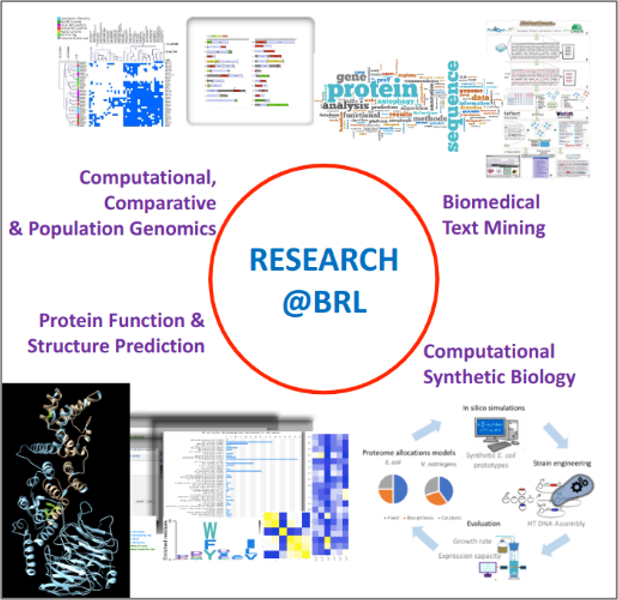
BiomedicalText MiningThe BRL actively develops machine learning methods and tools for biomedical text mining for concept discovery (Papanikolaou et al., 2011 & 2014; Theodosiou et al., 2024). Such tools can have general applicability across the biomedical sciences; they are made freely available to the research community as user-friendly web-apps and/or as source code, for customised use and expansion. We also develop custom, machine-learning-based applications that help our in-house biomedical literature curation efforts. A set of biomedical literature analysis tools were instrumental in the development of the LIRcentral database (Chatzichristofi et al., 2023) and are routinely used to enhance our biocuration work for the expansion of this unique resource. |
Our research interests focus on the development of empirical, statistical, machine-/deep-learning methods and specialized (curated) databases for exploiting available types of biological information towards understanding biological systems at the molecular, cellular and phenotypic level. More specifically, we devise algorithms, methods/tools, databases and computer systems for:
Computational Comparative GenomicsWith a particular interest in protein bioinformatics, the BRL has contributed significantly to the study of large classes of non-globular proteins: (a) We pioneered the development of widely used software tools (CAST, PlaToLoCo) and databases (LCR-eXXXplorer) for detecting and analysing protein sequences with local compositional bias (Promponas et al., 2000; Kirmitzoglou and Promponas, 2015) and integrated such modules in comparative genomics pipelines (Vasileiou et al., 2023). All these computational resources are freely accessible to the research community either as user-friendly web servers or as source code. (b) We performed systematic (Mier et al., 2020) and large-scale (Ntountoumi et al., 2019; Kastano et al., 2022) analyses to characterise classes of compositional bias and to suggest their possible functional roles and identify underlying evolutionary trends. (c) We exposed (and proposed solutions for) systematic errors present in current publicly available protein sequence databases stemming from either propagation of erroneous annotations (Promponas et al., 2015) or due to long protein repeats (Tørresen et al., 2019).
Case study:Computational Methods for studying Selective MacroautophagyDuring the last decade, our group has pioneered the development of computational approaches for studying key proteins involved in selective macroautophagy as cargo receptors, adaptors/scaffolds, via their interaction with Atg8 proteins mediated by a short linear peptide (LIR/AIM motif). |
(d) We were also the first to provide solid genomic and transcriptomic evidence for the presence and possible functionality of (almost intact) oligosaccharyl-transferase complexes in the malarial causing parasites of the genus Plasmodium (Tamana and Promponas, 2019). This discovery has revamped the international interest on the possible roles of N-linked glycosylation in the lifecycle (and possibly in the pathogenicity) of these parasites. Protein sequence, Function, Structure & EvolutionThe BRL has established a strong research direction towards the characterization and study of proteins and processes related to eukaryotic endomembrane systems (e.g., autophagy, nuclear pore complexes) with computational The BRL develops tools for characterizing nuclear pore complex subunits and their unconventional interactions within eukaryotic genomes (Katsani et al., 2014), established their evolutionary relations with other eukaryotic endomembrane systems (Promponas et al., 2016) and developed pipelines for fast & sensitive detection of fungal NUPs in vast metagenomic datasets (Nicolaides et al., 2023). BRL is leading the development of computational resources for studying selective autophagy, including the online iLIR prediction server (Kalvari et al, 2014) and iLIR Database (Jacomin et al, 2016), widely used by autophagy researchers worldwide for studying LIR motif-containing proteins (LIRCPs) across species. We have recently developed the LIRcentral resource (Chatzichristofi et al., 2023; https://lircental.eu), offering the richest collection of LIRCPs with experimentally verified LIRs, manually curated from the literature. LIRcentral curators participated in annotating the “Autophagy-related proteins” thematic dataset published with the most recent release of DisProt (https://www.disprot.org/; Aspromonte et al., 2024) and we currently exploit this rich dataset to develop novel, AI-based methods for predicting functional LIR motifs within eukaryotic proteomes and beyond.
We initially worked towards refining sequence-based descriptors for automatically detecting functional (i.e., binding) LIR motifs within sequences. This highly efficient set of computer tools (iLIR; Kalvari et al, 2014) is easily applicable in large sequence datasets (e.g., complete proteomes). Through the last decade, iLIR has been used for the discovery of dozens of new functional motifs from international research groups. With the application of iLIR in a set of model proteomes (including the human proteome), we could map for the first time a potential complete set of selective autophagy receptors/ adaptors/ scaffolds, making this information available in the web accessible iLIR Database (Jacomin et al., 2016). We currently work on the development, maintenance and expansion of the widest collection of LIR motifs supported from experimental data, manually curated from the biomedical literature. We now offer the LIRcentral database (Chatzichristofi et al., 2023) as a reference point for researchers across the globe who need to find all experimentally supported information on this topic. With such a high-quality dataset in our hand, we currently develop advanced machine learning methods for the enhanced prediction of functional LIR motifs. |
|
SELECTED GRANTS
|
SELECTED PUBLICATIONS
|

